using CSV, DataFrames, Transformers, Flux, Random, ProgressMeter
amino_codon = Dict( # the amino acid to codon relationship
"A" => ["GCU", "GCC", "GCA", "GCG"],
"R" => ["CGU", "CGC", "CGA", "CGG", "AGA", "AGG"],
"N" => ["AAU", "AAC"],
"D" => ["GAU", "GAC"],
"B" => ["AAU", "AAC", "GAU", "GAC"],
"Q" => ["CAA", "CAG"],
"E" => ["GAA", "GAG"],
"Z" => ["CAA", "CAG", "GAA", "GAG"],
"G" => ["GGU", "GGC", "GGA", "GGG"],
"H" => ["CAU", "CAC"],
"I" => ["AUU", "AUC", "AUA"],
"L" => ["CUU", "CUC", "CUA", "CUG", "UUA", "UUG"],
"K" => ["AAA", "AAG"],
"M" => ["AUG"],
"F" => ["UUU", "UUC"],
"P" => ["CCU", "CCC", "CCA", "CCG"],
"S" => ["UCU", "UCC", "UCA", "UCG", "AGU", "AGC"],
"T" => ["ACU", "ACC", "ACA", "ACG"],
"W" => ["UGG"],
"Y" => ["UAU", "UAC"],
"V" => ["GUU", "GUC", "GUA", "GUG"],
#"1" => ["AUG"],
#"9" => ["UAA", "UGA", "UAG"],
);Transformers
Modern Sequence Analysis
Traditional Sequence Analysis
Sequence analysis has been typically performed by recurrent neural networks.
Exploding/vanishing gradients from recursion.
Information decay leading to short memory.
O(n) complexity in sequence length.
Some solutions.
LSTMs: forget gates to preserve information.
Gated Recurrent Units: similar to LSTM forget gates but no output.
Other proposals: none ideal due to O(n) complexity.
Transformers
“Attention is all you need” (2016)
Transformers are the latest development in large scale sequence analysis.
Address many problems with RNNs
Workhorse behind many “magical” applications e.g. voice assistant and language translation.
Attention: bare bones
Transformers leverage the idea of attention: not new.
Attention is computed between all elements in a sequence: a weighted relationship between locations.
All units are considered independently: massive parallelisation.
Attention
Simple attention relies on nothing more than a Euclidean projection: dot product.
![]()
Simple attention: computation
- For each vector pair compute the dot product between them and normalise over all pairs with
softmax. \[ a_{ij} = x_i^T x_j \] \[ A_{ij} = \frac{\exp(a_{ij})}{\sum_j \exp(a)_{ij}} \]
Attention Retrevial
An input is weighted by the row of the attention matrix corresponding to its index.
The ouput is the weighted sum of all vectors by this attention row: \[ y_i = \sum_j A_{ij} x_j \]
Querys, Keys, and Values
Imagine the vectors are one-hot batched: \[(0,0,1, \ldots, 0,0)\]
The product \(x_i^Tx_j\) will be one only in the indexes \(i=j\). Multiplying by the potential valus gives \(x_i\).
This operation is acting like a look-up table.
Transformers inherit the language and call these keys, queries, and values.
Attention Generalised
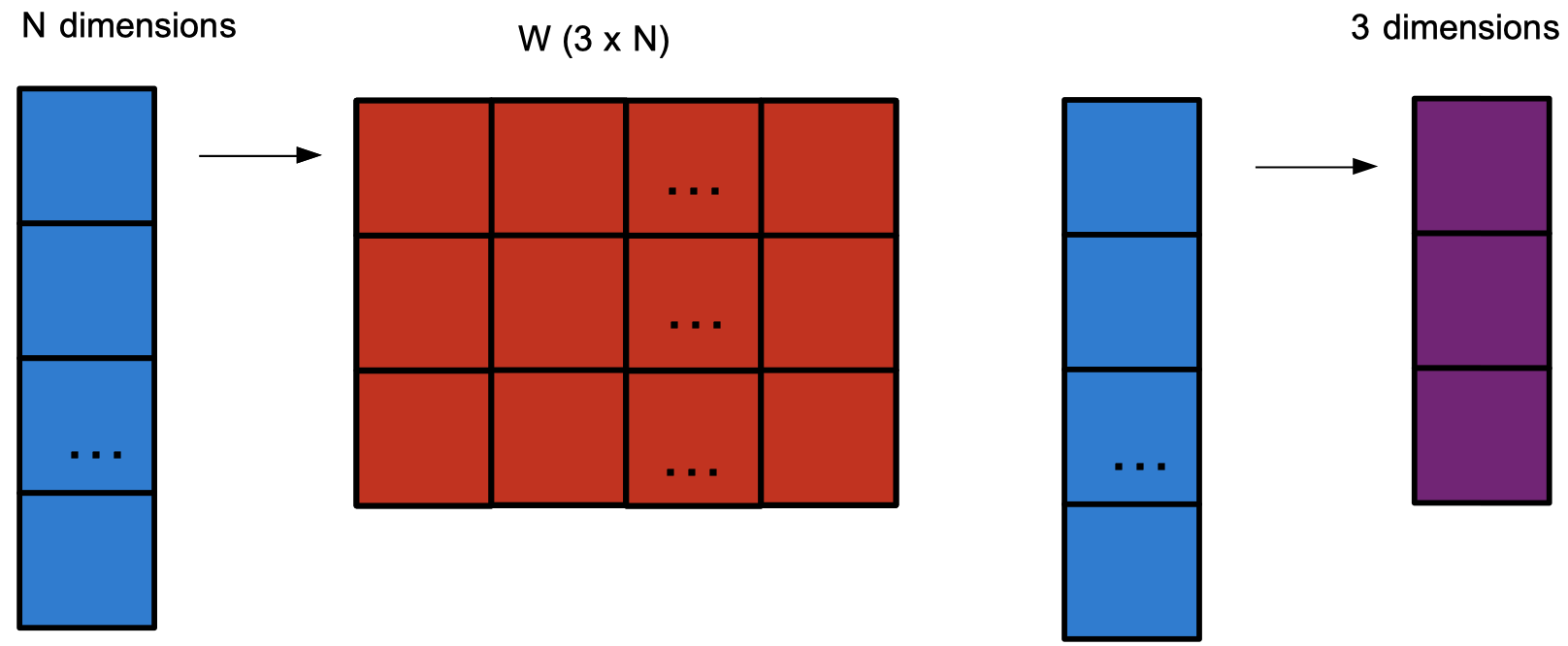
We would like to let the keys, queries, and values not be fully determined by Euclidean representation.
We imagine them as linear transforms of the total dictionary embedding values into a new space: \[k_i = W^Kx_i, q_i = W^Qx_i, v_i = W^Vx_i\]
This allows us to compress our key/query/value representation to a lower dimensionality.
Transformers Bare Bones
The transformer model is composed of two attentional models: an encoder and decoder.
Input is in the form of vectors of tokens e.g. genome sequence.
The encoder transforms input to an encoded attentional sequence.
The decoder autoregressively uses the encoder ouput to an ouput sequence.
The output sequence is decoded with a decoder dictionary e.g. amino acids.
Transformers.jl
Transformers.jlis built on top of Flux.It has similar GPU support (and similiar drawbacks).
Similar grammar but there are subtleties - documentation is helpful!
Open source contribution - documentation could be added to! Anyone can do this.
Transfomer Codon Task
A codon is a group of three base nucleotides that encodes an amino acid.
Can we try and predict amino acid base pairs without knowledge of codons?
Pre-Processing
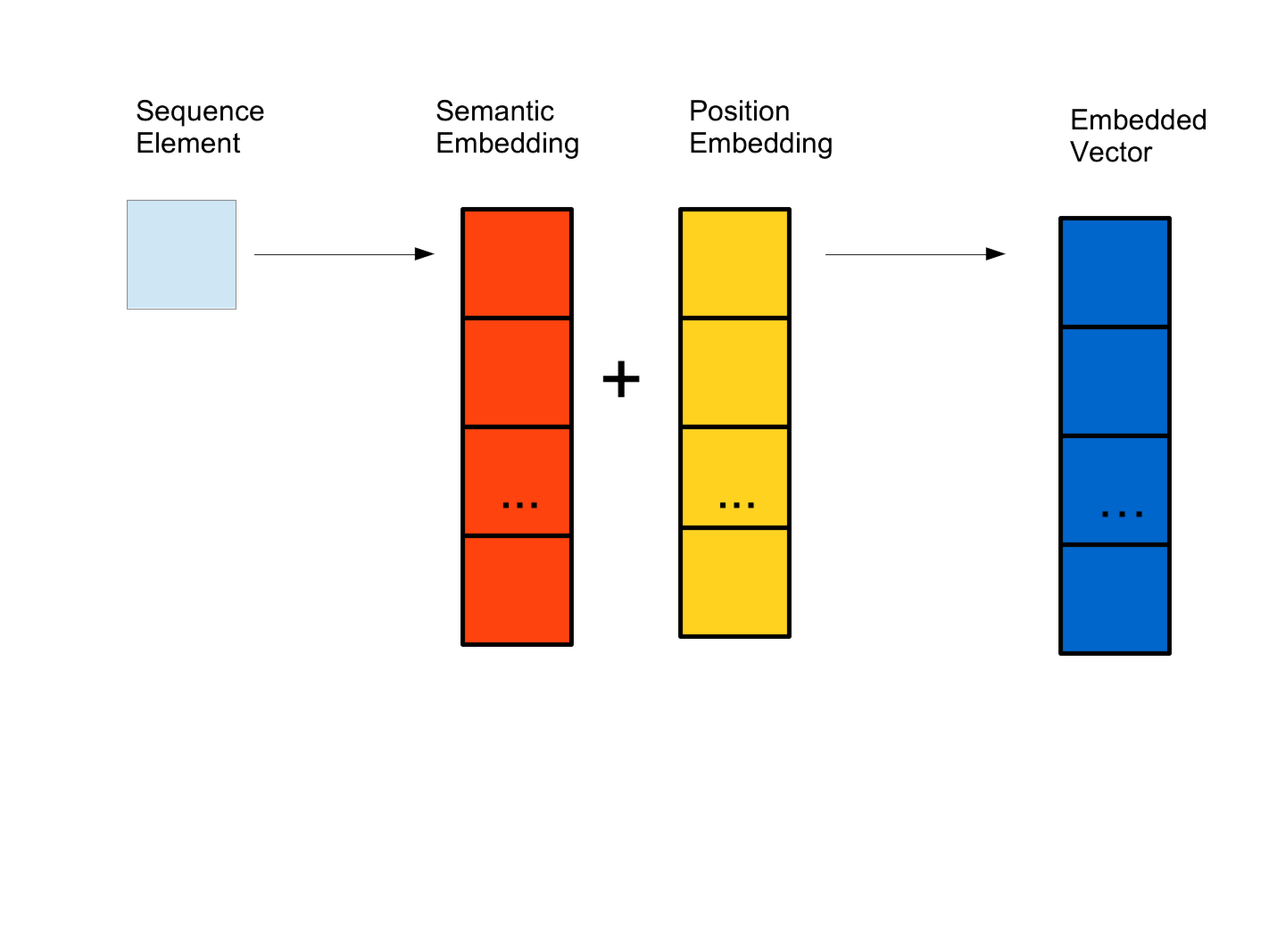
Token labels, a start/end symbol, and an unknown symbol are encoded into a lookup table (Vocabulary).
The input sequence tokens are first wrapped by “Start” and “End” tokens and encoded by the Vocabulary.
This encoded representation is semantically embedded into vectors of length \(D\).
pre_process(v) = cat("1", v..., "9"; dims=1)
labels_y = cat("1", unique(train_y[1])..., "9", "0"; dims=1)
labels_x = cat("1", unique(train_x[1])..., "9", "0"; dims=1)
tokenizer_x = Transformers.Vocabulary(labels_x, "0")
tokenizer_y = Transformers.Vocabulary(labels_y, "0")
encoded_x = tokenizer_x.(pre_process.(train_x))
encoded_y = tokenizer_y.(pre_process.(train_y))
embed_x = Transformers.Embed(d, length(tokenizer_x))
embed_y = Transformers.Embed(d, length(tokenizer_y))Positional Encoding
Attention lets us forget sequence order to parallelise computation.
Sequence orders are important e.g. gene sequence cant be scrambled.
A positional encoding function is used to inject this order into the learning.
- Positional encodings can be learnt but typically use a function: \[ p_{2k}(i) = cos(i/10000^k) \] \[ p_{2k+1}(i) = sin(i/10000^k) \]
Keys, Queries, and Values
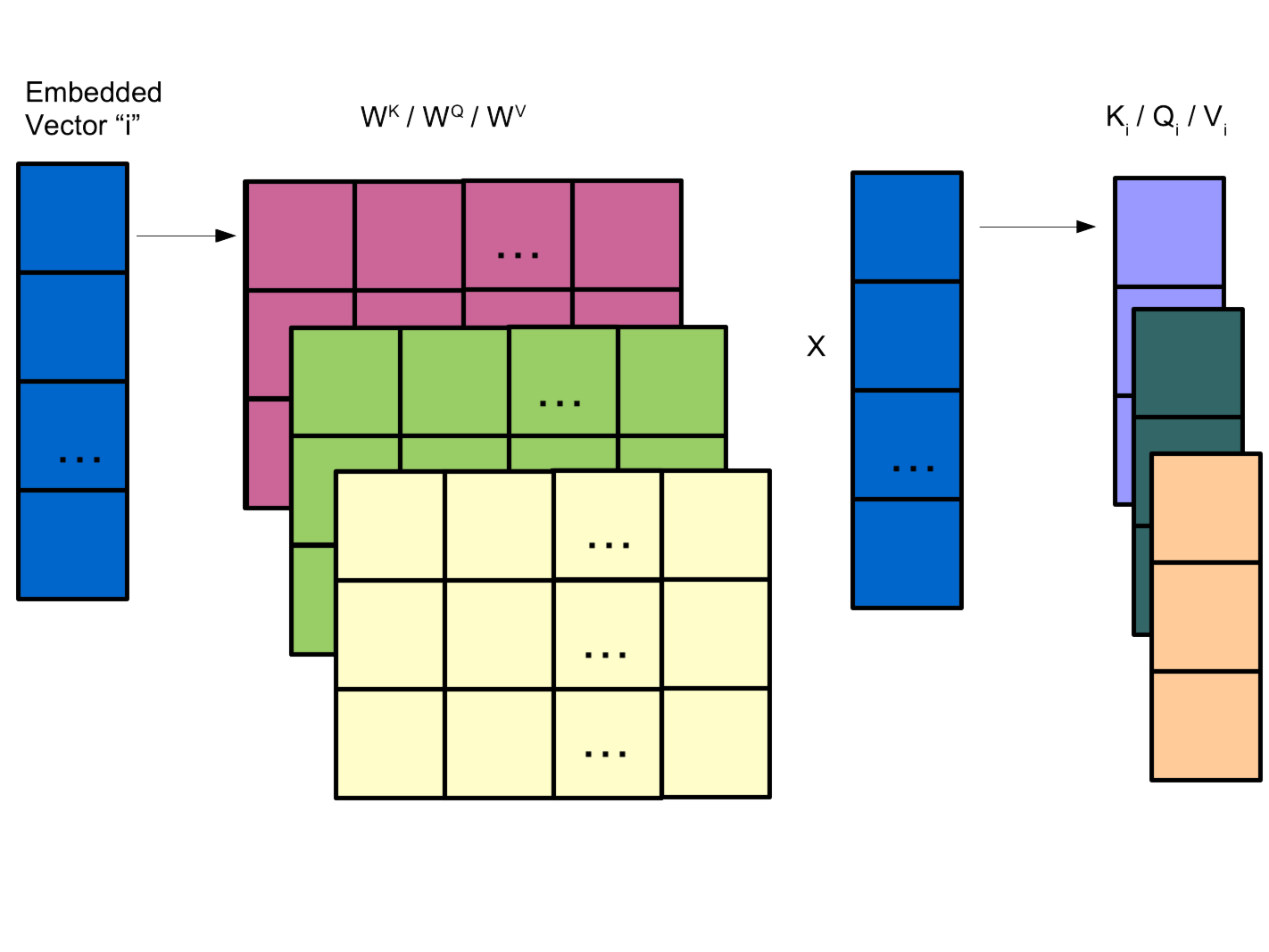
The keys, queries and values are remembeddings of the information into a different representational subspace.
These are the key learnable parameters in the model.
An efficient subspace representation of information allows for impressive generalisability.
Attention Matrices
Each input vector forms a row and these can be concatenated for efficiency into a matrix: \(X\) (L x D)
K represents the keys, Q the querys, V the values.
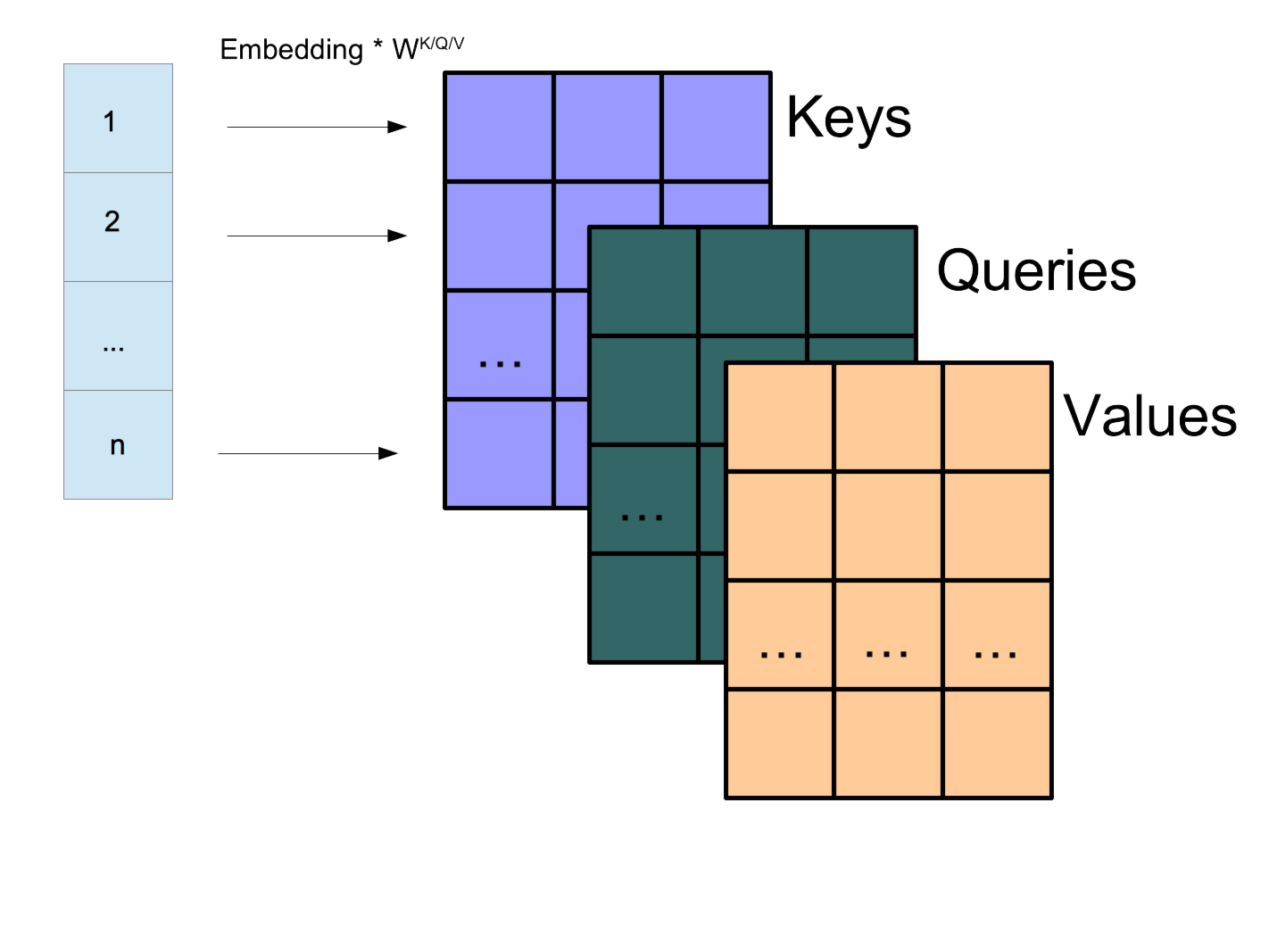
Transformer Attention Head
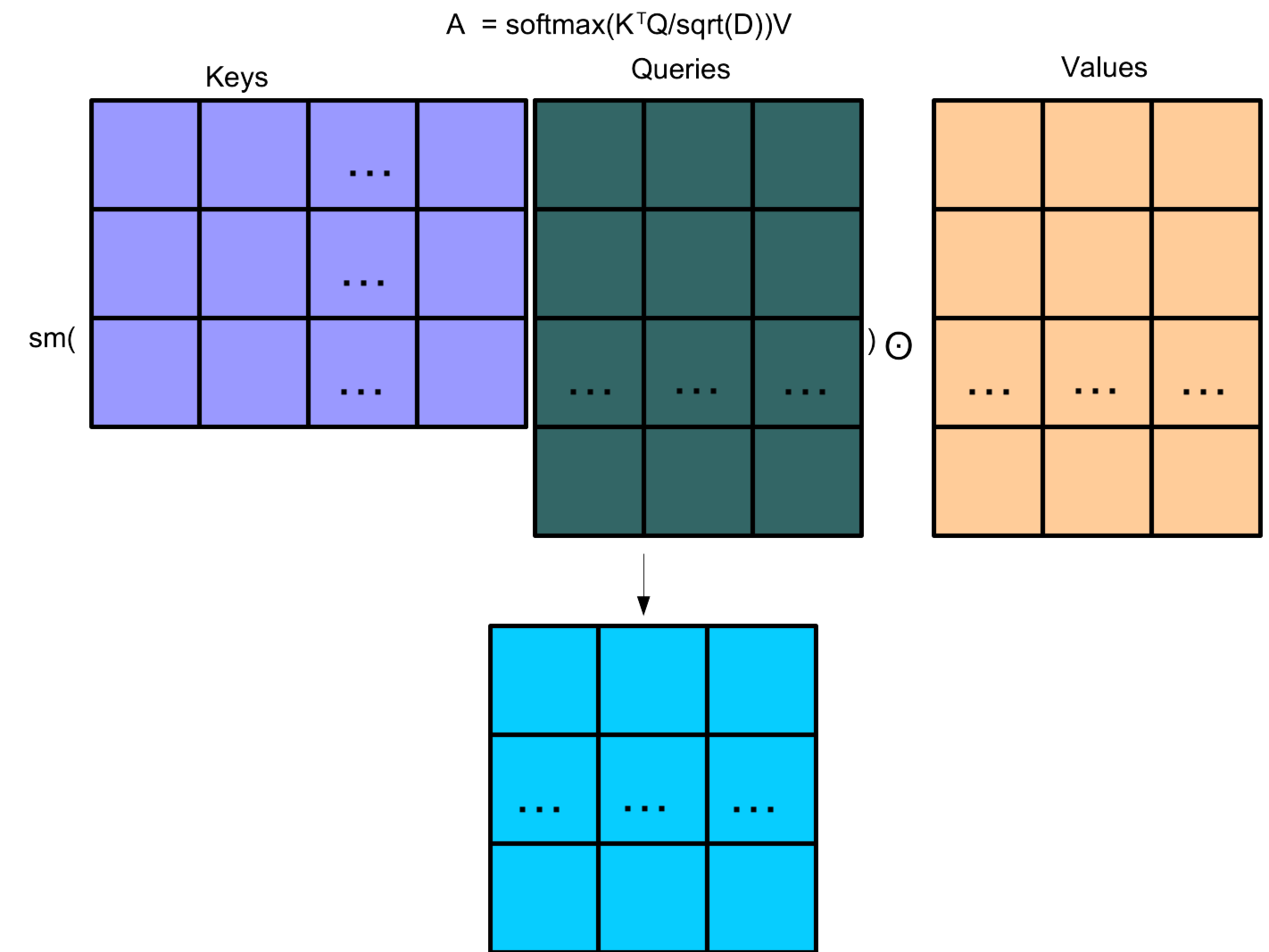
The Transformer attention mechanism is given by: \[A(K,Q,V) = \text{softmax}\left(\frac{K^TQ}{\sqrt{D}}\right) \odot V\] \[K = W^KX, Q = W^QX, V = W^VX\]
Softmax is calculated row by row. \(\odot\) is element-wise product. Dimensional scaling \(\sqrt(D)\) for stability.
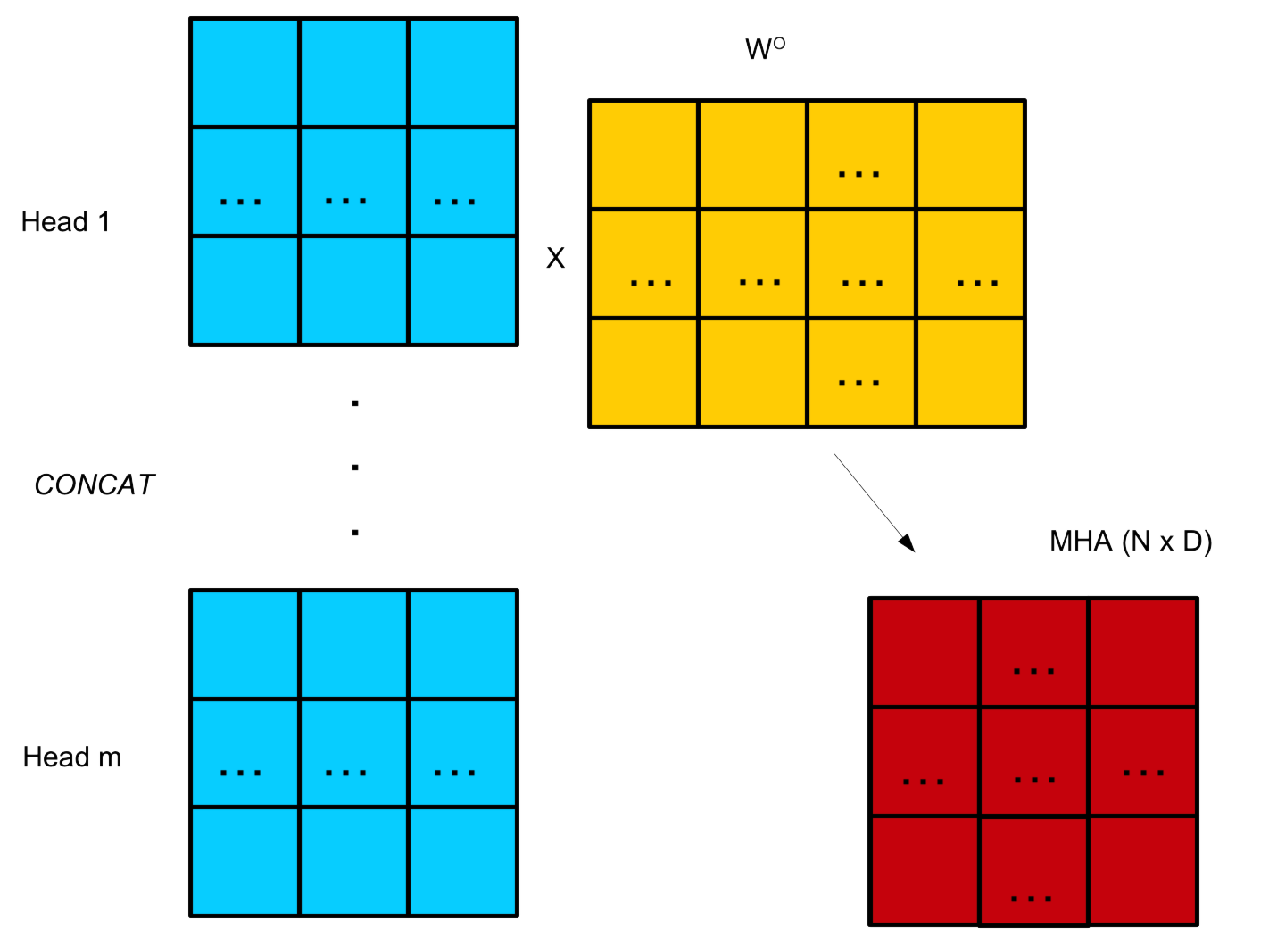
Multi-head attention
A transformer layer can have multiple heads analag. convolutional filter.
Each of these heads will learn to focus on different semantic relationships.
This can be efficiently encoded by simply concatenating each individual head. Usually, \(n \times h_d = D\).
Head dimension is not a hyper-parameter.
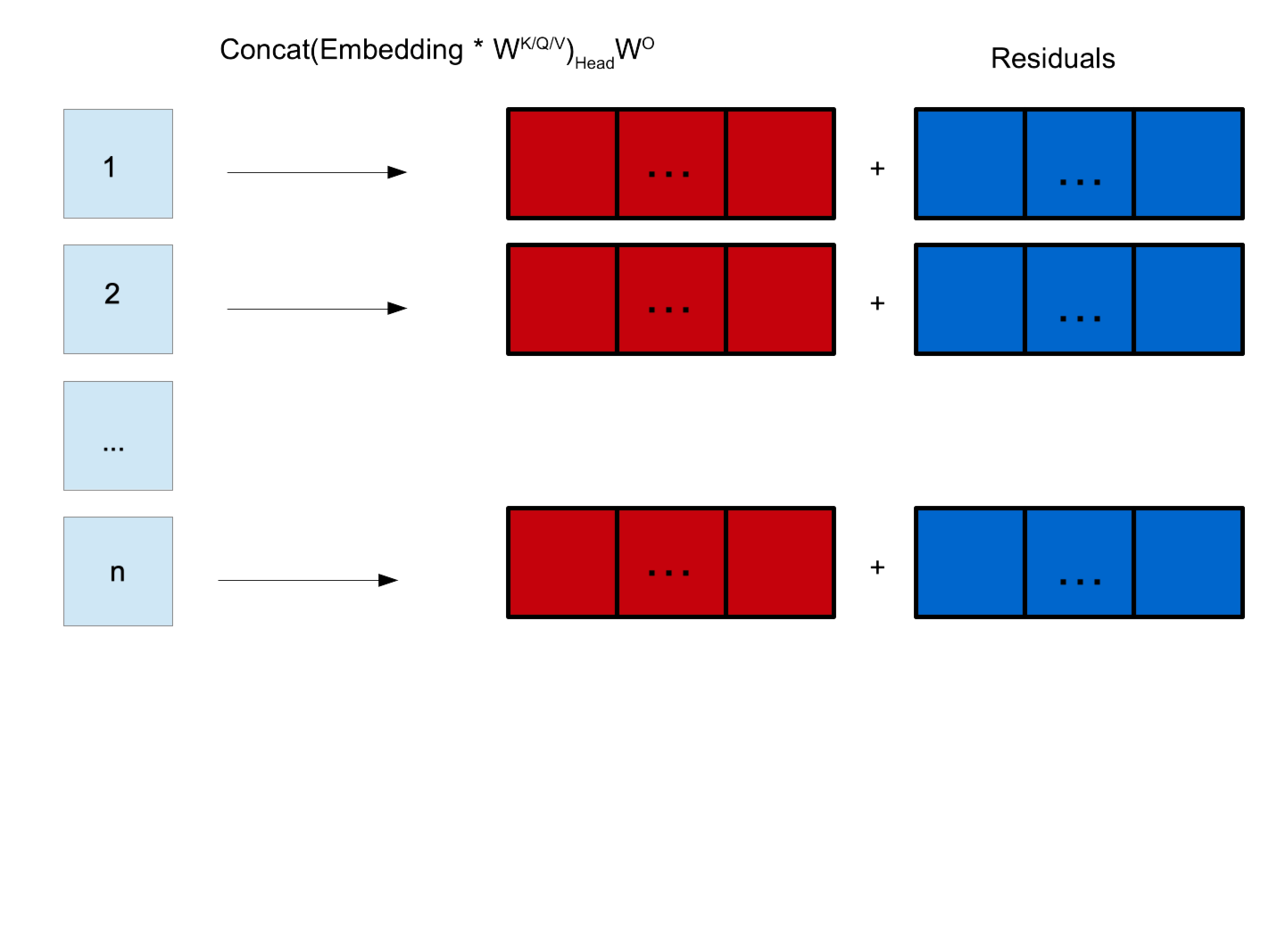
Residual and Normalisation
The outputs of the attention mechanism are mutliplied by matrix \(W^O\) to remebbed vectors into length D.
\(W^O\) is another learnable matrix.
The pre-attention inputs (residuals) are added to the re-embedded attention transformed inputs.
This combined vector is then layer-normalised.
Feed Forward Network
- The normalised self-attention and residuals are passed through a feed-foward network: \[F(x) = \text{ReLu}(W_1x + b_1)W_2 + b_2\]
Inner Dimensions
The inner-dimension is independent of the embedding dimension e.g. 2048.
The feed forward network is shared between all tokens.
The residuals are added to the ouput of the FFN and layer-normalised.
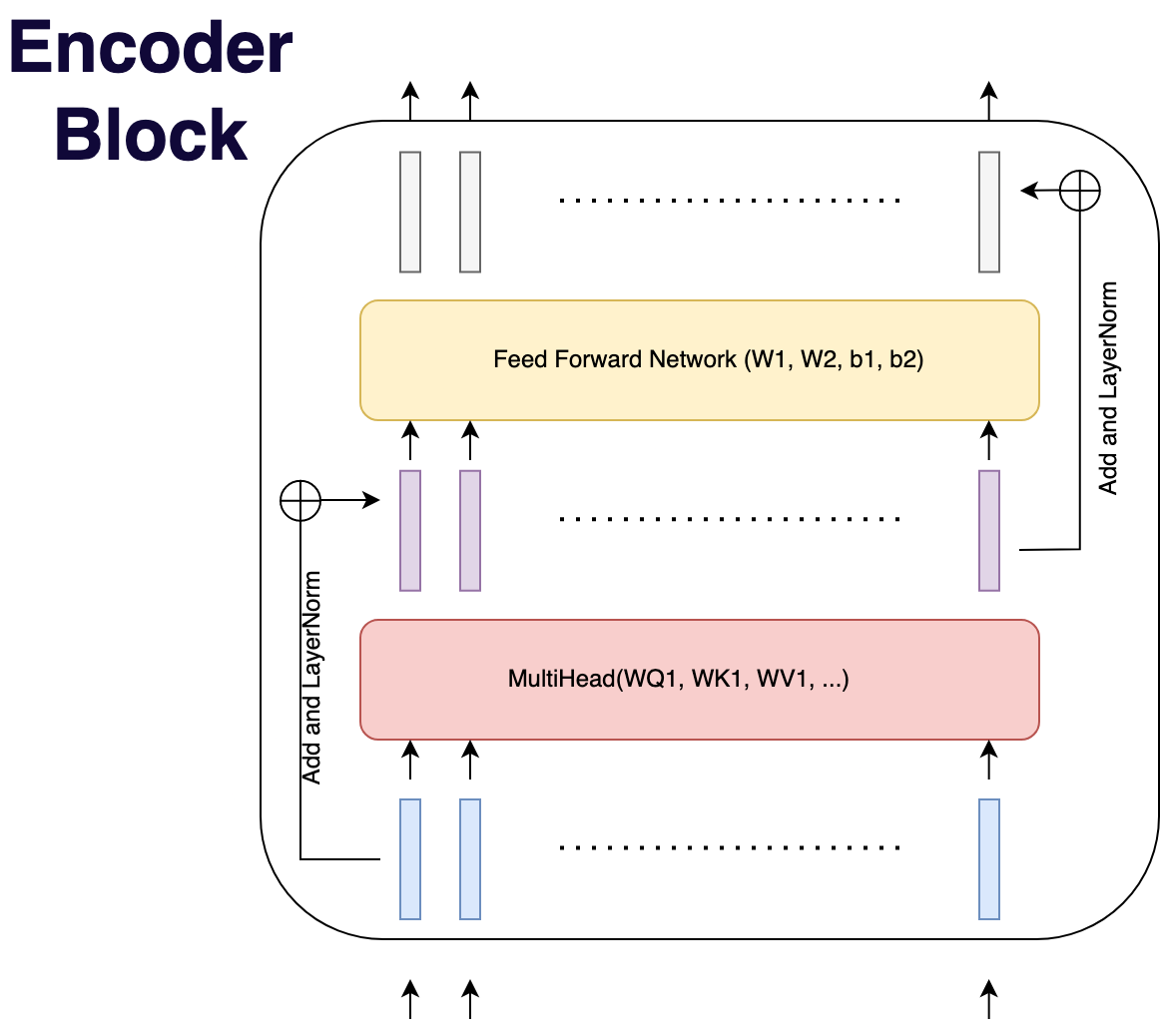
Encoder Block/Layer
The operations of self-attention, feed forward networks, and layer-normalisation make an encoder block.
This can be thought of as a layer in a regular NN.
The Encoder is formed of several encoder blocks e.g. 6
Decoder Block
A decoder block has a self-attention, an encoder-decoder-attention, and a feed-forward network.
The encoder-decoder-attention keys and values are constructed using output from encoder (and learnable matrices).
These keys and values are shared across all decoder blocks.
Dec1 = TransformerDecoder(d, h, hd, innerd)
Dec2 = TransformerDecoder(d, h, hd, innerd)
Dec3 = TransformerDecoder(d, h, hd, innerd)
ffn = Transformers.Positionwise(Dense(d, length(tokenizer_y)), softmax)
function Tdecoder((y, mx))
emy = embeddingy(y)
d1 = Dec1(emy, mx)
d2 = Dec1(d1, mx)
d3 = Dec1(d2, mx)
ffn(d3)
end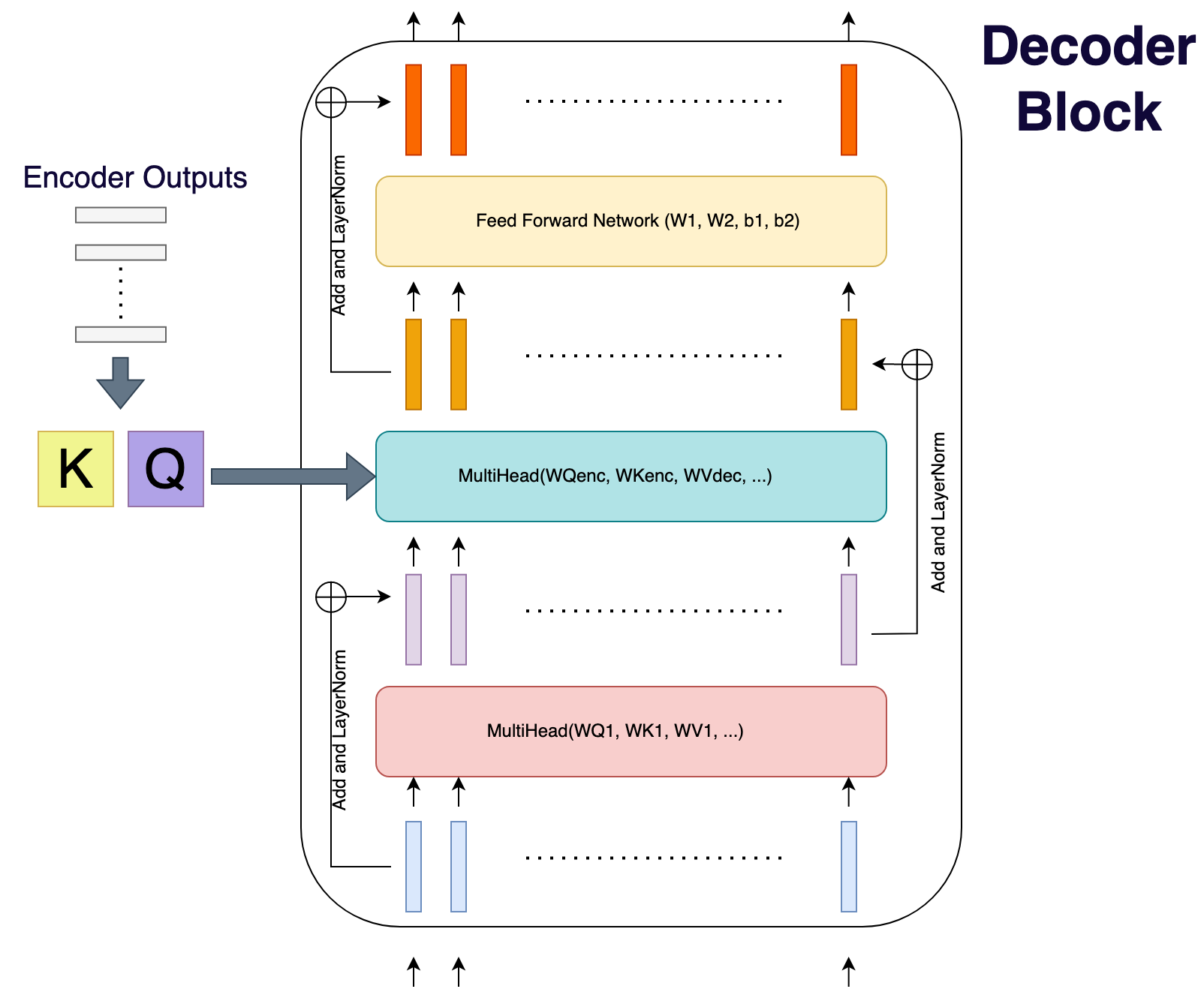
Sequence Generation
Sequences are generated with a start symbol and terminated with a stop symbol.
The outputs are fed through the network as inputs until stopping.
The output vector is fixed at an arbitrary length i.e. 1024
Future outputs are masked with
-Infto prevent left flowing information: autoregressive.
function transcribe_protein(x)
seq = [tokenizer_y("1")]
tok = ["1"]
enc = Tencoder(x)
for i = 1:2*length(x)
dec = Tdecoder((seq, enc))
seqnext = argmax(vec(dec[1:end-1,end]))
append!(seq, seqnext)
toknext = Transformers.decode(tokenizer_y, seqnext)
push!(tok, toknext)
toknext == "9" && break
end
tok
endLoss Functions
The final step is to
softmaxoutputs to generate a probability distribution against a dictionary/vocabulary.The natural loss function is
crossentropy.Loss functions may be arbitrary.
Regularisation
The original paper used dropout for the layer parameters and label smoothing.
Dropout improves stability and convergence time.
Label smoothing increases model perplexity (at the cost of labelled accuracy).

Model Summary
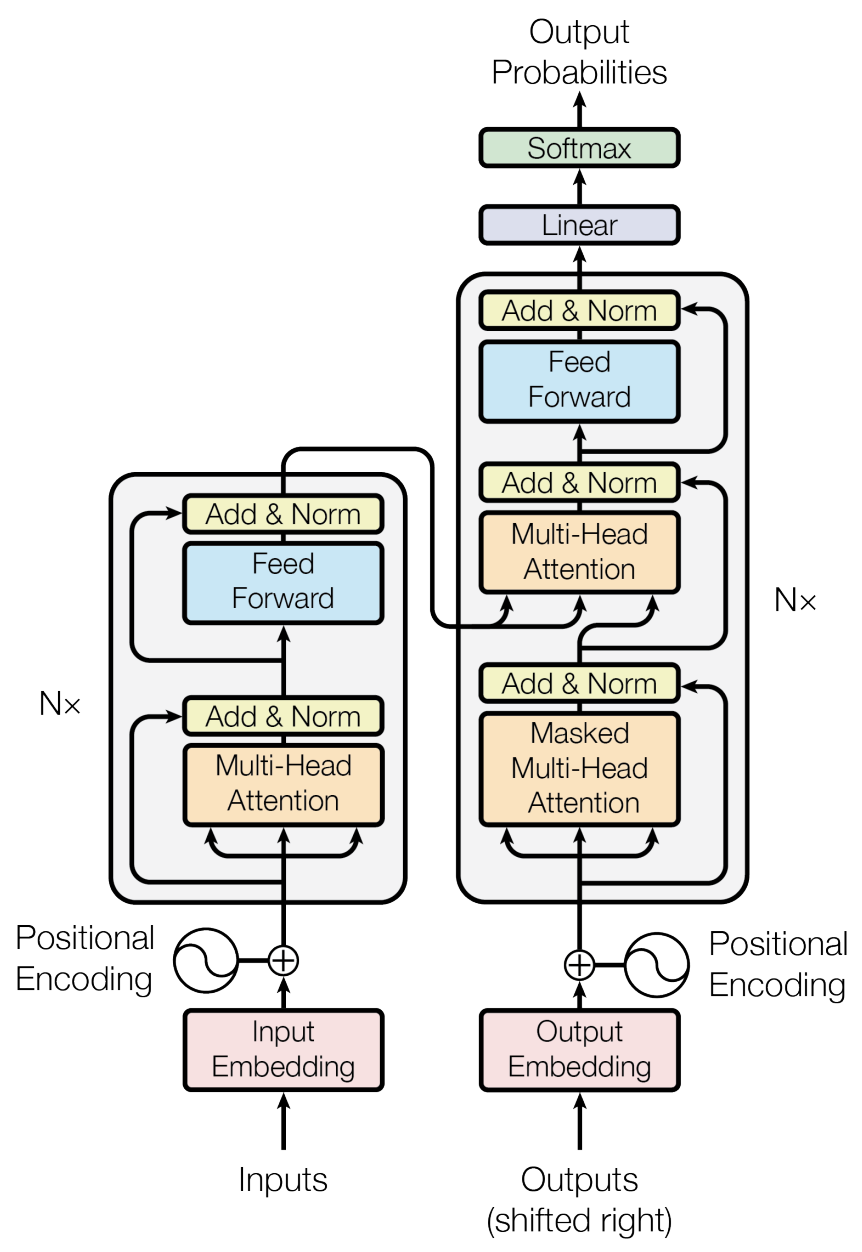
The encoder takes positional and contextual inputs and the decoder autoregressively produces outputs.
The encoder and decoders use attention heads and a feed-forward network to perform the learning.
The attention mechanism transforms embedding into a different subspace through keys/queries/values matrices.
Keys, Queries, Values are learned and represent optimal information relationships in the problem context.
Transfer Learning
Transformer models are large - amongst the largest neural networks.
Difficult to train without industrial resources.
Employ transfer learning: import pre-trained weights on a similar task and fine-tune to current task.
Transformers in Biology
Transformers are in relative infancy - lots of work to be done.
The obvious candidate is sequence analysis: genome and protein.
Some interesting developments: gene transcription factors using Enformer (Deep Mind)
Protein Prediction tasks.
Transforming the Language Of Life.
Protein prediction can be done classically: HMMs and BLAST. Exponentially computationally expensive.
CNNs and RNNs are computationally more efficient but task depedent and dont generalise.
Authors propose Transformer model PRoBERTa: pre-trained agnostic amino acid sequence representation.
Protein prediction tasks: binary PPI and protein family classification.
Problems
Authors managed to achieve state of the art performance on target tasks (tasks not too important)
The resulting model is vastly more computationally efficient: 128 GPUs for 4 days => 4 GPUs for 18 hours.
Still difficult to reproduce. GPUs are top of the line and few people have access to this many.
Transformers in general are large models and pose a reproducibility problem.
Summary
Transformers are a complex but powerful sequence analysis architecture.
Julia offers support through
Transformers.jl.Very successful, but computationally demanding and not easily reproducible.
Relatively unplumbed in Biology.
References
Attention is all you need; Vaswani et. al. (2017)